Introduction
In this guide, we’re going to implement the linear support vector machine algorithm from scratch in Python. Our goal will be to minimize the cost function, which we’ll use to train our model, and maximize the margin, which we’ll use to predict values against new, untrained data. We’ll be using NumPy — one of Python’s most popular machine learning libraries — to handle all of our calculations, so you won’t need any additional software or libraries installed on your computer to follow along! If you want to have a quick overview of the basics of SVM and its calculations check this article: Primal Formulation of SVM: A Simplified Guide | Machine Learning.
The main goal of SVM
We looked at the working of SVM in detail in previous articles, but to give a quick understanding of the goal of SVM let's explain it! The main goal of SVM is the maximization of margins between two different classes. That means that you want to make sure that as many points in one class are on one side of the decision boundary and as many points in the other class are on the other side.
When this happens, all points with a higher degree of separation will be correctly classified while all points with a lower degree of separation will be misclassified.
.png) |
| SVM Diagram |
So when the marginal distance between these two classes is at their maximum, they become the optimal solution for maximizing margin and minimizing risk. As such, it becomes a lot easier to classify new points without any error because they can just be placed on either side of the decision boundary based on which class it belongs to. If there are errors though then there's always something called regularization which allows us to penalize models so that they generalize better for new data points.
The equation for Soft Margin SVM
Implementing SVM from scratch in python
Writing the SVM class
# svm.pyimport numpy as npclass SVM:def __init__(self, C = 1.0):# C = error termself.C = Cself.w = 0self.b = 0
Hinge Loss calculation
# Hinge Loss Function / Calculationdef hingeloss(self, w, b, x, y):# Regularizer termreg = 0.5 * (w * w)for i in range(x.shape[0]):# Optimization termopt_term = y[i] * ((np.dot(w, x[i])) + b)# calculating lossloss = reg + self.C * max(0, 1-opt_term)return loss[0][0]
Gradient Descent optimization
def fit(self, X, Y, batch_size=100, learning_rate=0.001, epochs=1000):# The number of features in Xnumber_of_features = X.shape[1]# The number of Samples in Xnumber_of_samples = X.shape[0]c = self.C# Creating ids from 0 to number_of_samples - 1ids = np.arange(number_of_samples)# Shuffling the samples randomlynp.random.shuffle(ids)# creating an array of zerosw = np.zeros((1, number_of_features))b = 0losses = []# Gradient Descent logicfor i in range(epochs):# Calculating the Hinge Lossl = self.hingeloss(w, b, X, Y)# Appending all losseslosses.append(l)# Starting from 0 to the number of samples with batch_size as intervalfor batch_initial in range(0, number_of_samples, batch_size):gradw = 0gradb = 0for j in range(batch_initial, batch_initial + batch_size):if j < number_of_samples:x = ids[j]ti = Y[x] * (np.dot(w, X[x].T) + b)if ti > 1:gradw += 0gradb += 0else:# Calculating the gradients#w.r.t wgradw += c * Y[x] * X[x]# w.r.t bgradb += c * Y[x]# Updating weights and biasw = w - learning_rate * w + learning_rate * gradwb = b + learning_rate * gradbself.w = wself.b = breturn self.w, self.b, losses
Predict Method
def predict(self, X):prediction = np.dot(X, self.w[0]) + self.b # w.x + breturn np.sign(prediction)
Performing Classification
Creating random dataset
# prediction.pyfrom sklearn import datasetsimport matplotlib.pyplot as pltimport numpy as npfrom sklearn.metrics import accuracy_scorefrom sklearn.model_selection import train_test_splitfrom svm import SVM# Creating datasetX, y = datasets.make_blobs(n_samples = 100, # Number of samplesn_features = 2, # Featurescenters = 2,cluster_std = 1,random_state=40)# Classes 1 and -1y = np.where(y == 0, -1, 1)
Splitting the dataset
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.5, random_state=42)
Training the SVM model
svm = SVM()w, b, losses = svm.fit(X_train, y_train)
Making predictions and testing accuracy
prediction = svm.predict(X_test)# Loss valuelss = losses.pop()print("Loss:", lss)print("Prediction:", prediction)print("Accuracy:", accuracy_score(prediction, y_test))print("w, b:", [w, b])----Loss: 0.0991126738798482Prediction: [-1. 1. -1. -1. 1. 1. 1. 1. -1. 1. -1. -1. 1. 1. -1. -1. 1. -1.1. -1. 1. 1. -1. 1. 1. 1. -1. -1. -1. -1. 1. -1. -1. 1. 1. -1.1. -1. -1. 1. 1. 1. 1. 1. 1. -1. 1. 1. 1. 1.]Accuracy: 1.0w, b: [array([[0.44477983, 0.15109913]]), 0.05700000000000004]
Visualizing SVM
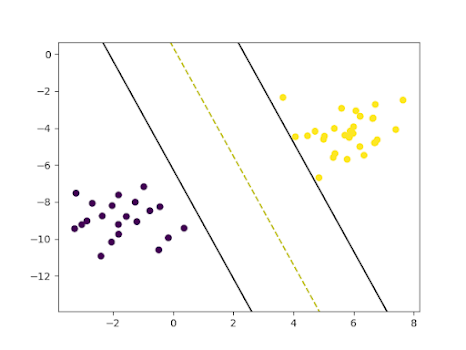
# Visualizing the scatter plot of the datasetdef visualize_dataset():plt.scatter(X[:, 0], X[:, 1], c=y)# Visualizing SVMdef visualize_svm():def get_hyperplane_value(x, w, b, offset):return (-w[0][0] * x + b + offset) / w[0][1]fig = plt.figure()ax = fig.add_subplot(1,1,1)plt.scatter(X_test[:, 0], X_test[:, 1], marker="o", c=y_test)x0_1 = np.amin(X_test[:, 0])x0_2 = np.amax(X_test[:, 0])x1_1 = get_hyperplane_value(x0_1, w, b, 0)x1_2 = get_hyperplane_value(x0_2, w, b, 0)x1_1_m = get_hyperplane_value(x0_1, w, b, -1)x1_2_m = get_hyperplane_value(x0_2, w, b, -1)x1_1_p = get_hyperplane_value(x0_1, w, b, 1)x1_2_p = get_hyperplane_value(x0_2, w, b, 1)ax.plot([x0_1, x0_2], [x1_1, x1_2], "y--")ax.plot([x0_1, x0_2], [x1_1_m, x1_2_m], "k")ax.plot([x0_1, x0_2], [x1_1_p, x1_2_p], "k")x1_min = np.amin(X[:, 1])x1_max = np.amax(X[:, 1])ax.set_ylim([x1_min - 3, x1_max + 3])plt.show()visualize_dataset()visualize_svm()
Visualization of dataset
Visualization of SVM on the test set
Complete version of SVM algorithm
import numpy as npclass SVM:def __init__(self, C = 1.0):# C = error termself.C = Cself.w = 0self.b = 0# Hinge Loss Function / Calculationdef hingeloss(self, w, b, x, y):# Regularizer termreg = 0.5 * (w * w)for i in range(x.shape[0]):# Optimization termopt_term = y[i] * ((np.dot(w, x[i])) + b)# calculating lossloss = reg + self.C * max(0, 1-opt_term)return loss[0][0]def fit(self, X, Y, batch_size=100, learning_rate=0.001, epochs=1000):# The number of features in Xnumber_of_features = X.shape[1]# The number of Samples in Xnumber_of_samples = X.shape[0]c = self.C# Creating ids from 0 to number_of_samples - 1ids = np.arange(number_of_samples)# Shuffling the samples randomlynp.random.shuffle(ids)# creating an array of zerosw = np.zeros((1, number_of_features))b = 0losses = []# Gradient Descent logicfor i in range(epochs):# Calculating the Hinge Lossl = self.hingeloss(w, b, X, Y)# Appending all losseslosses.append(l)# Starting from 0 to the number of samples with batch_size as intervalfor batch_initial in range(0, number_of_samples, batch_size):gradw = 0gradb = 0for j in range(batch_initial, batch_initial+ batch_size):if j < number_of_samples:x = ids[j]ti = Y[x] * (np.dot(w, X[x].T) + b)if ti > 1:gradw += 0gradb += 0else:# Calculating the gradients#w.r.t wgradw += c * Y[x] * X[x]# w.r.t bgradb += c * Y[x]# Updating weights and biasw = w - learning_rate * w + learning_rate * gradwb = b + learning_rate * gradbself.w = wself.b = breturn self.w, self.b, lossesdef predict(self, X):prediction = np.dot(X, self.w[0]) + self.b # w.x + breturn np.sign(prediction)

.png)

.png)
.png)
.png)